Island biogeography
island-biogeo.RmdThe theory of Island Biogeography, first articulated by Robert Macarthur and E.O. Wilson in the 1964 paper “Equilibrium theory of insular zoogeography”, presents a quantitative and predictive approach to understanding biodiversity on island systems. This framework assumes that species immigrate at higher rates from the mainland to nearby islands than to islands further away, and that extinction rates are higher in smaller islands than larger islands. The balance of these two processes determine an island’s equilibrium species richness. Macarthur and Wilson expanded on this theory in their classic 1967 book “The Theory of Island Biogeography”.
There are many ways to specifically turn this descriptive framework into a mathematical model – for example, we can model a linear relationship between island size and extinction rates, or we can imagine it as a nonlinear relationship. In this package we assume the following nonlinear relationships between island size and extinction rates, and between distance and immigration rates:
Extinction rate on island:
\[E = e^{\frac{kS}{A}}-1\]
Immigration rate from mainland to island: \[I = e^{\left(-\frac{k}{D}*(S-M)\right)}-1\]
This results in an equilibrium number of species on the island as
follows:
\[S_{eq} = \frac{AM}{D+A}\]
where:
-
\(D\) is the distance from the
mainland to the island (km);
-
\(A\) is the area of the island
(km^2);
-
\(M\) is the number of species on
the mainland;
- \(k\) is the scaling constant (we set to \(k=0.015\) in this implementation, but the exact number is not very important).
We can simulate the dynamics of this model in R using
the function run_ibiogeo_model(). This function runs the
isoland biogeography model for two islands at a time, and we need to
provide as inputs the size of the two islands and their distance from
the mainland:
distances <- c(1, 2) # Distance of the two islands from mainland
areas <- c(2,2) # size (area) of the islands
ibiogeo_out <- run_ibiogeo_model(D = distances, A = areas)The output from run_ibiogeo_model() is two plots – one
of which is a diagram of a map showing the two islands, and the other
which shows the immigration/extinction rates and resulting equilibrium
species richness for the two islands:
ibiogeo_out$map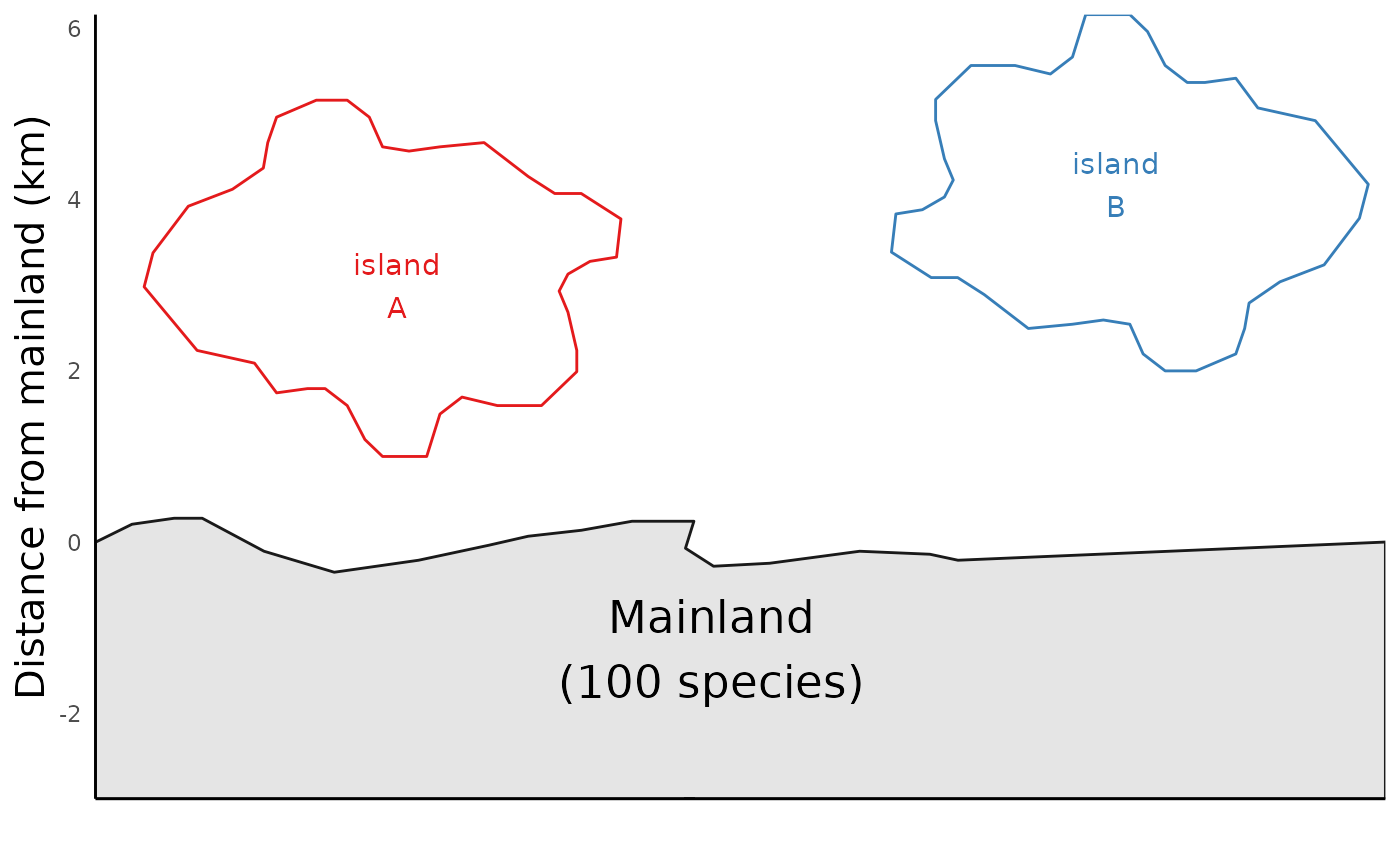
ibiogeo_out$eq_plot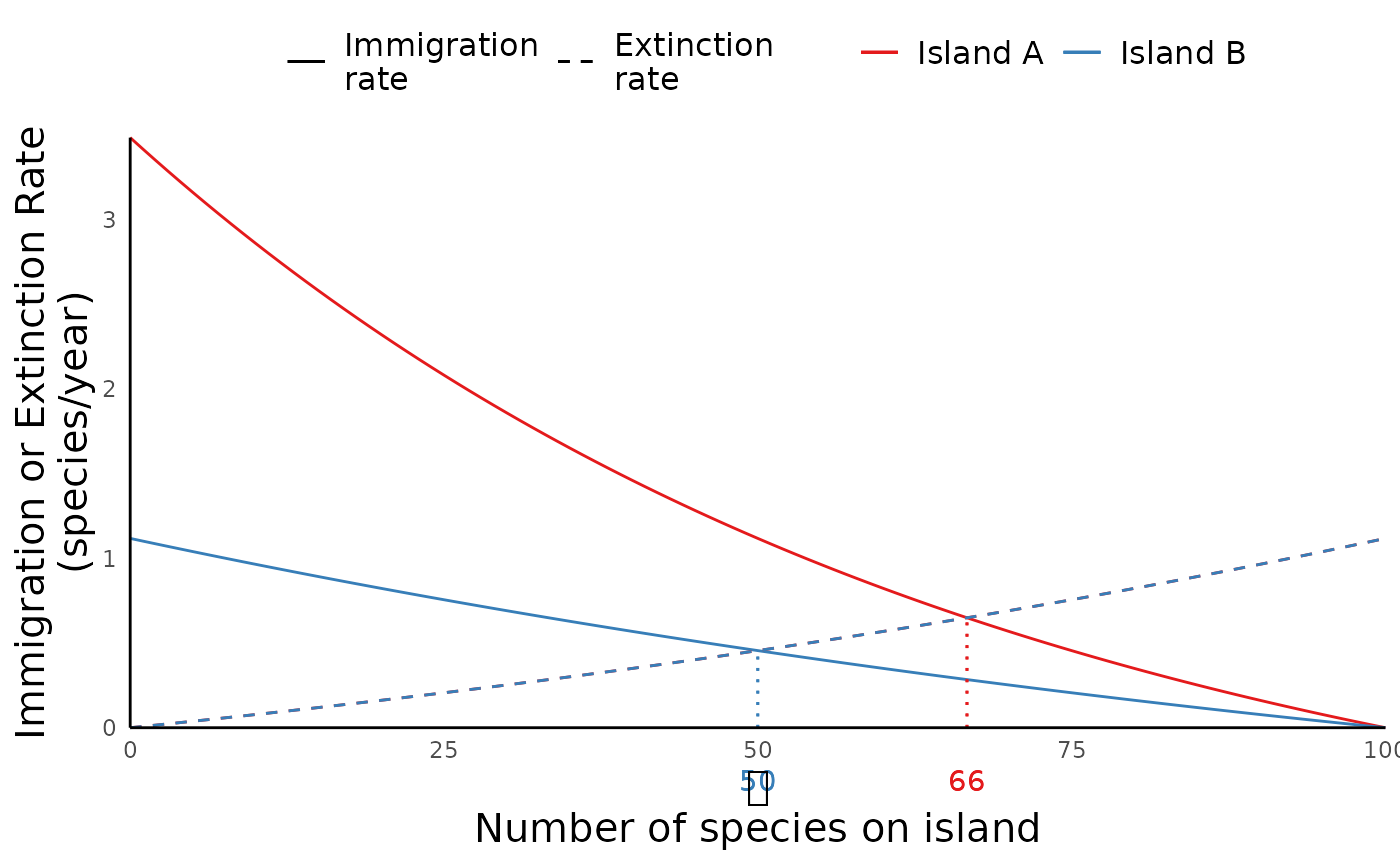
In this case, the extinction rate curve for both islands are same, since we set island sizes to be identical. This is why we just see the one line.
As we see in the map plots above, run_ibiogeo_model()
assumes by default that the mainland is home to 100 species. But we can
change this parameter as well:
mainland_richness <- 50
distances2 <- c(1, 2) # Distance of the two islands from mainland
areas2 <- c(2,1) # size (area) of the islands
ibiogeo_out2 <- run_ibiogeo_model(D = distances2, A = areas2, M = mainland_richness)
ibiogeo_out2$map 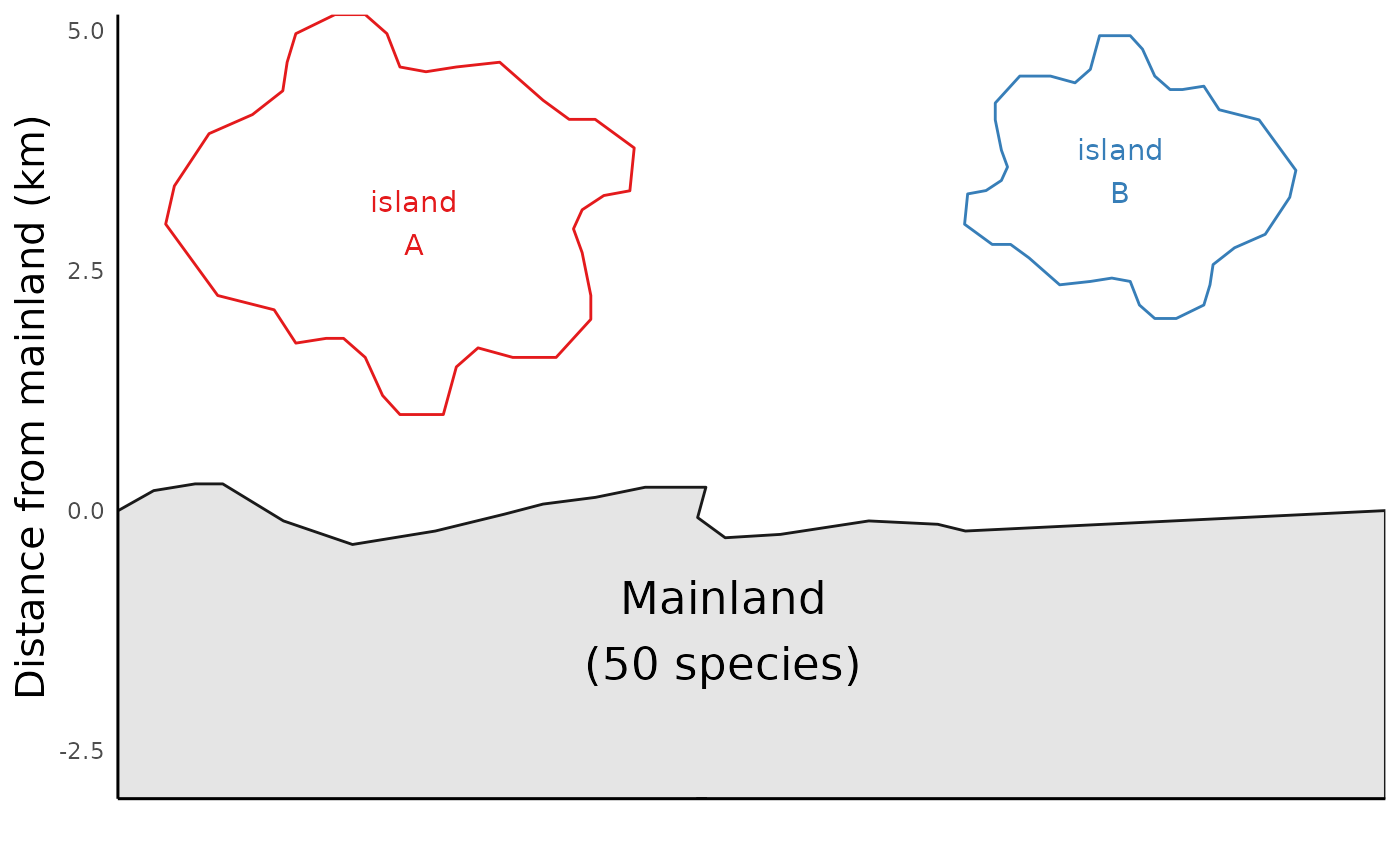
ibiogeo_out2$eq_plot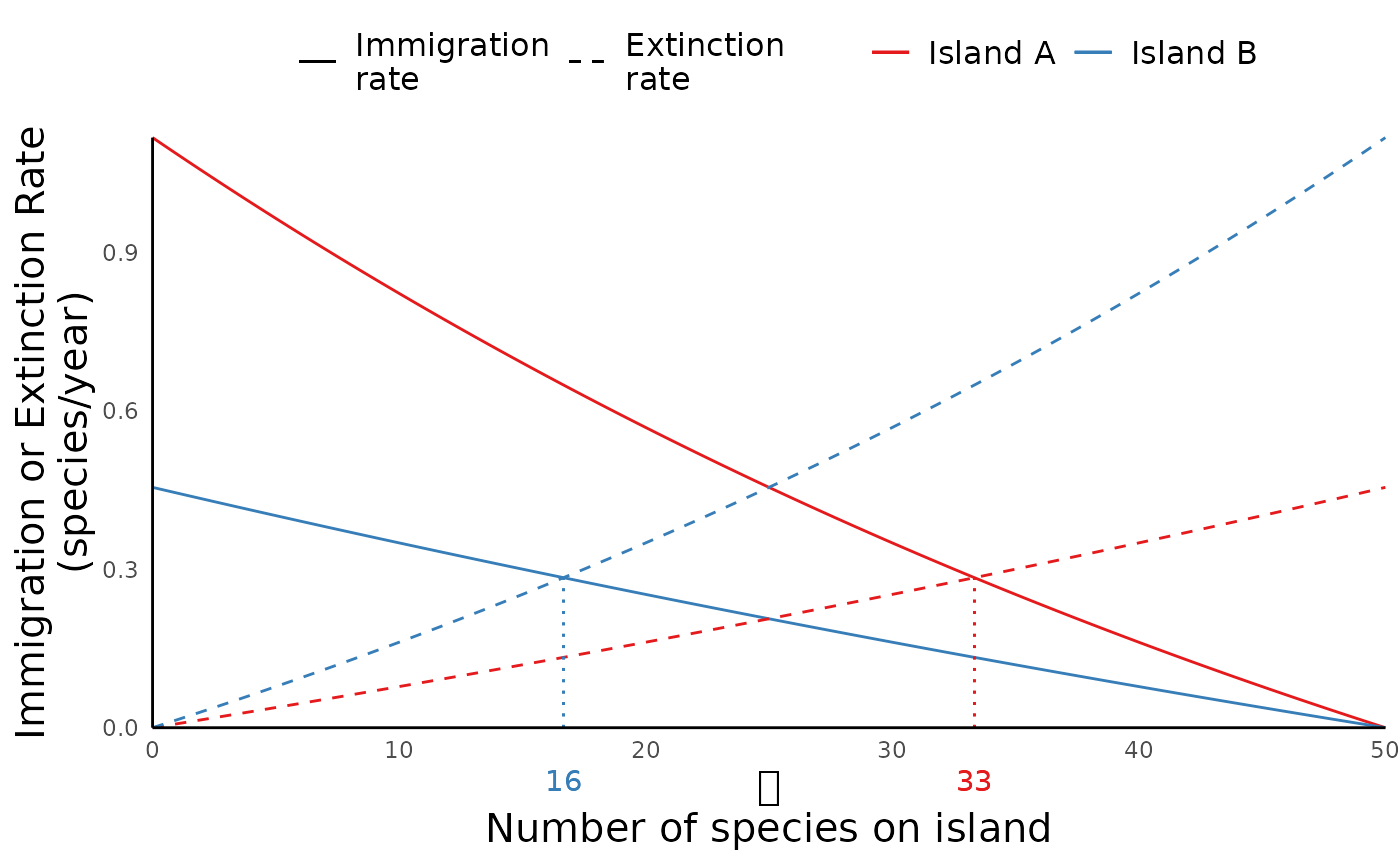
Further reading
Daniel Simberloff notably conducted experimental tests of this theory using mangrove islands in Florida, and island biogeography has also been hugely influential in conservation biology. For a recent review of island biogeography’s influence, see this 2017 review.